Research Projects
-

HIV Host Interactions
-
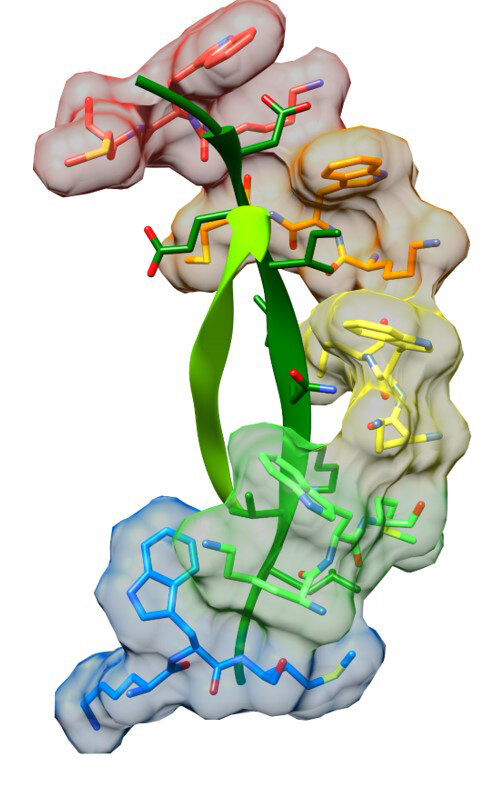
AAA Protein Translocases
-

D-Peptide Therapeutics
-

Chromatin Regulation
-

Insulin Therapeutics
-

Ubiquitin-Proteasome System
HIV-host Interactions
We are partners in the HIV P50 HIV structural biology consortium – CHEETAH (Center for the Structural Biology of Cellular Host Elements in Egress, Trafficking, and Assembly of HIV). Our interests and contributions include architectural components of the virion and cellular factors that either counteract the virus or are recruited to facilitate viral replication.
AAA Protein Translocases
AAA ATPases comprise a large family of protein machines that perform much of the mechanical work in cells. A large class of these enzymes, known as the AAA unfoldases, form hexamers that translocate their protein substrates through the central pore. This typically unfolds the substrate to disassemble a protein complex or enable degradation, although the many family members perform a multitude of biological functions. Our structural and biochemical studies, which are collaborative with Peter Shen and Wes Sundquist indicate that AAA unfoldases use a sequential rotary mechanism to translocate and unfold their substrates.
D-peptide therapeutics
Peptides have potential as therapeutics because they have the potential to block protein-protein interactions and are relatively easy to synthesize and optimize. Unfortunately, they are not generally suitable for clinical use because they are susceptible to degradation by naturally occurring protease, which can also contribute to immunogenicity. We are collaborating with Michael Kay, who is developing peptides made from D-amino acids, which have the opposite (mirror-image) chirality but otherwise identical properties to their naturally-occurring counterparts. Current targets include inhibition of a conformational change of the envelope protein that is needed for HIV virus to enter a host cell and an analogous approach to inhibition of the ebola virus.
Chromatin Regulation
In collaboration with Tim Formosa, we are investigating the biochemical, structural, and functional basis for molecular interactions that enable and regulate chromatin biology. This includes studying the structure and function of histone chaperones, such as FACT and Spt6, which enable the assembly, disassembly, and reorganization of nucleosomes. We are also investigating the network of interactions that these proteins make with their functional binding partners, such as RNA polymerase.
Insulin Therapeutics
Although insulin therapeutics are widely used in the treatment of diabetes, limitations on the currently available molecules include susceptibility to aggregation, modest response rates (even for current “fast-acting” insulins, and lack of dynamic response to glucose levels. In an effort to develop novel insulin therapeutic lead compounds that display improved properties, we are collaborating with Danny Chou. A first success in this program has been the development of a four-disulfide insulin that retains biological activity, maintains native insulin structure, and is resistant to aggregation. This advance may inform efforts to develop a therapeutic that can be used in the absence of cold-chain delivery and as long-term insulin pump devices.
Ubiquitin-Proteasome System
We have a long-standing interest in regulation of the proteasome, which is the major protease of the cytosol and nucleus. This includes determining a series of structures of complexes that revealed the mechanism of regulating access to the central compartmentalized protease. We also study enzymes and protein-interactions of the ubiquitin system, in which the addition or removal of the small protein ubiquitin from other target proteins often regulates target degradation by the proteasome and has a profound impact on many facets of cellular function.
